Modelling the SIMPLE model of memory
Bayesian cognitive modelling
In this blogpost I am going to use, yet, another example from the book “Bayesian Cognitive Modelling” by Lee & Wagemakers. This book is full of examples in cognitive psychology which can be approached in a Bayesian way. Since the examples are mostly done via WinBUGS, I wanted to see if I can recreate it using brms and stan. So, that is what this blogpost will be about. (I know the book also provides the stan codes, but I want to see how far I can get with brms which has less functionality then base STAN or WinBUGS).
In this example, I will model the SIMPLE model of memory, which measures people’s ability to retain stimuli. The challenge of this model, as you will come to discover, is within its nonlinear curve.
The background is not that important, but what it is the way on how to approach the problem. Let me show you by plotting the data.
But first, load the libraries and the data.
# clears workspace:
rm(list=ls())
library(dplyr)
library(ggplot2)
library(brms)
library(bayesplot)
library(tidybayes)
library(marginaleffects)y <- matrix(scan("CaseStudies/Simple/k_M.txt", sep=","), ncol=40, nrow=6, byrow=T)
n <- c(1440,1280,1520,1520,1200,1280)
listlength <- c(10,15,20,20,30,40)
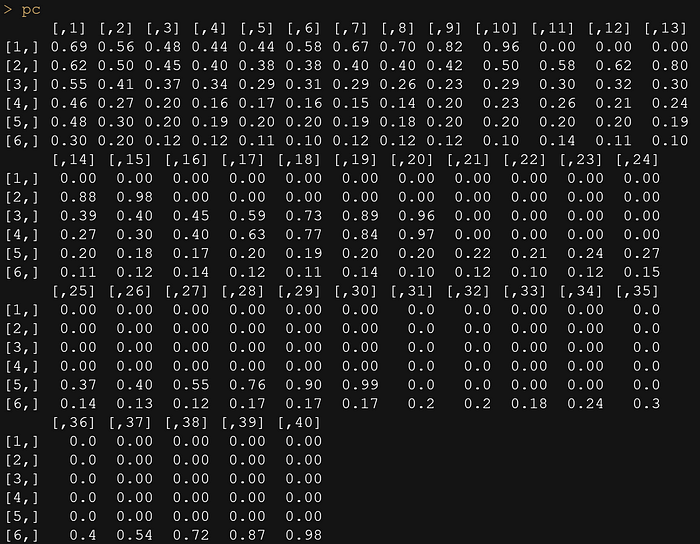
pc <- matrix(scan("CaseStudies/Simple/pc_M.txt", sep=","), ncol=40, nrow=6, byrow=T)
labs <- scan("CaseStudies/Simple/labs_M.txt", sep="", what="character")
dsets <- 6
gsets <- 9
y
pc
labs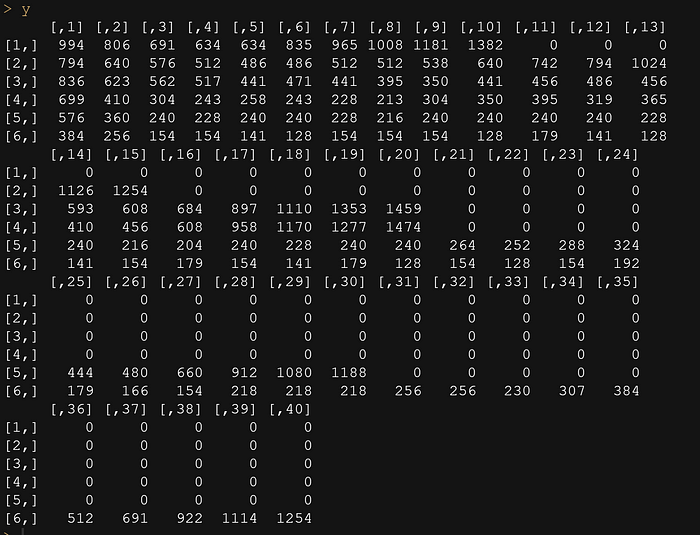

pc<-as.data.frame(t(pc))
colnames(pc)<-labs
d <- pc
Position <- rownames(d)
rownames(d) <- NULL
df <- cbind(Position,d)
str(df)
df_long<-df%>%
tidyr::pivot_longer(cols=`10-2`:`40-1`,
names_to = "Condition",
values_to = "PC")
str(df_long)
df_long$Position<-as.numeric(df_long$Position)
df_long<-df_long%>%filter(PC>0)
ggplot(df_long)+
geom_point(aes(x=Position,
y=PC))+
theme_bw()+
facet_wrap(~Condition)
ggplot(df_long)+
geom_point(aes(x=Position,
y=PC))+
geom_smooth(aes(x=Position,
y=PC))+
theme_bw()+
facet_wrap(~Condition)

In the book, it is clearly show how complex the model is, also from a cognitive point of view. The models that are then most efficient to use are mechanistic models in which the various parts of the underlying theory are connected to form a model and from which to draw predictions. That model is also what is made in WinBUGS in the book and which can be created using STAN, but I would like to model it using basic brms. Now, this does not mean brms is basic, but I will do a slighlty different model than what is being done in the book. In the end, my job is to model the curve.
I can show the same graph from above using a long format. For that I will need to do some additional transformations.
y<-as.data.frame(t(y))
colnames(y)<-labs
d <- y
Position <- rownames(d)
rownames(d) <- NULL
df <- cbind(Position,d)
str(df)
df_long<-df%>%
tidyr::pivot_longer(cols=`10-2`:`40-1`,
names_to = "Condition",
values_to = "Y")
df_long$Position<-as.numeric(df_long$Position)
df_long<-df_long%>%
arrange(Condition, Position)
df_long$N<-rep(n,each=40)
df_long$PC<-df_long$Y/df_long$N
df_long<-df_long%>%filter(PC>0)
ggplot(df_long)+
geom_point(aes(x=Position,
y=PC))+
geom_smooth(aes(x=Position,
y=PC))+
theme_bw()+
facet_wrap(~Condition)

Now, the data is by all means non-linear and there are multiple ways of modelling it. We could use splines, a non-linear function, or we could transform the data and then model it via a more simple solution. Lets try the last suggestion first.
ggplot(df_long)+
geom_point(aes(x=log10(Position),
y=log10(Y)))+
geom_smooth(aes(x=log(Position),
y=log10(Y)))+
theme_bw()+
facet_wrap(~Condition)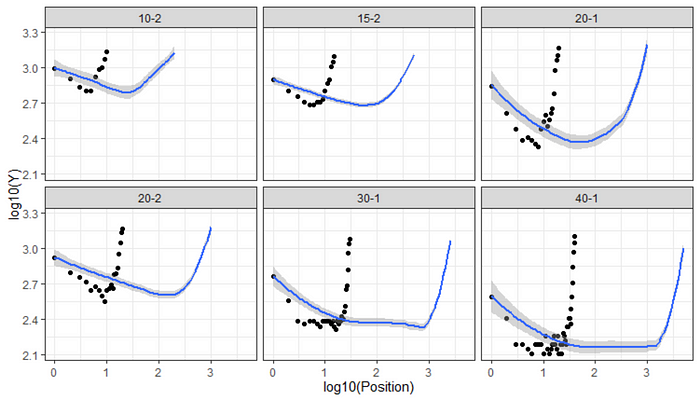
Of to the modelling part. Now, in a previous example I already showed that using the underlying Binomial distribution is a better choice than using the Beta distribution. So lets go for Binomial from the start.
get_prior(Y | trials(N) ~
Position + (1|Condition),
data = df_long,
family=binomial(link = "logit"))
fit<-brms::brm(Y | trials(N) ~
Position + (1|Condition),
data = df_long,
family=binomial(link = "logit"),
prior = c(
prior(normal(0, 1), class=b, coef=Position ),
prior(normal(0, 1), class=Intercept),
prior(gamma(1, 1), class=sd, coef=Intercept, group=Condition)),
chains=4,
cores=6,
warmup = 3000,
iter = 6000)
summary(fit)
plot(fit)
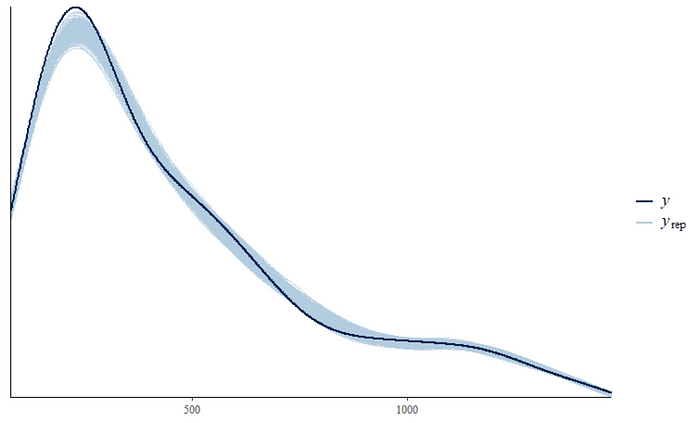
pp_check(fit, ndraws = 500)
pp_check(fit, type = "error_hist", ndraws = 11)
pp_check(fit, type = "scatter_avg", ndraws = 100)
pp_check(fit, type = "stat_2d")
pp_check(fit, type = "loo_pit")
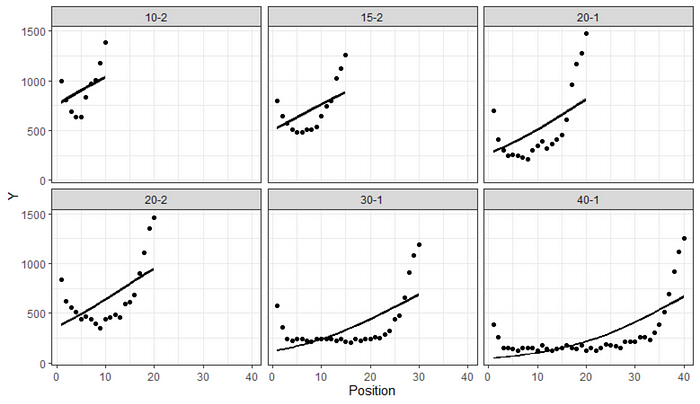
df_long %>%
add_epred_draws(fit,
ndraws=100,
allow_new_levels = TRUE) %>%
ggplot(aes(x = Position,
y = Y,
group=Condition)) +
geom_line(aes(y = .epred, group = paste(Condition, .draw)), alpha = 0.25) +
geom_point(data = df_long, color="black")+
facet_wrap(~Condition)+
theme_bw()


The predictions do not connect to the observations and when you look at the deviations, one would be very hard pressed to maintain that chosen priors and models are sufficient. I would not bet on that.
Another try then, but this time using splines. Splines are very nice way of modelling non-linear patterns, which are still linear in their function. However, the trouble is that a spline often looses part of its interpretation, especially when compared to a non-linear function which is harder to model.
Lets apply the splines in the brms model and see where we end up. The gp part of the spline constitutes a Gaussian Process. Many different types of splines are possible.
fit4<-brms::brm(Y | trials(N) ~
1 + s(Position, bs="gp") + (1+Position|Condition),
data = df_long,
family=binomial(link = "logit"),
chains=4,
cores=6,
warmup = 3000,
iter = 6000)
summary(fit4)
pp_check(fit4, ndraws=500)
df_long %>%
add_epred_draws(fit4,
ndraws=100,
allow_new_levels = TRUE) %>%
ggplot(aes(x = Position,
y = Y,
group=Condition)) +
geom_line(aes(y = .epred, group = paste(Condition, .draw)), alpha = 0.25) +
geom_point(data = df_long, color="black")+
facet_wrap(~Condition)+
theme_bw()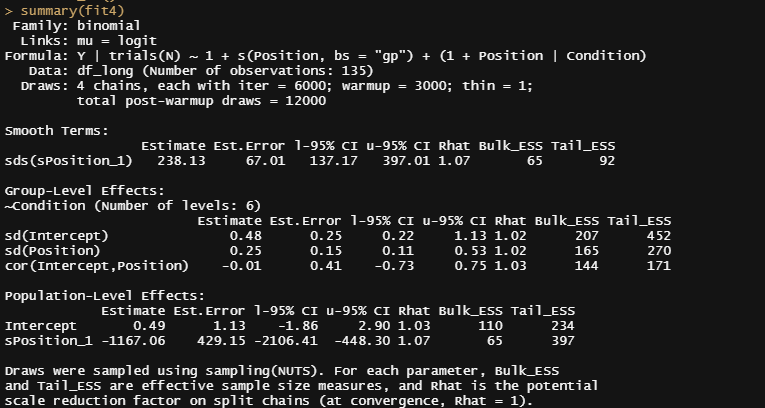

Hence, the spline is not the best way to fit the data. There are of course other methods which I will explore now, of which implementing a non-linear function is the best possible way.
Non-linear functions are notorious difficult, so I will start small using a single condition.
df_long_group2<-df_long%>%filter(Condition=="20-1")
plot(df_long_group2$Position, df_long_group2$Y)
Next up I want to apply the non-linear function using the nls program. The formula I am using now is one I found when looking for which to model a parabola. The sinusoidal function should not surprise you as it can model the ups and downs of a cycle, but I wonder if it will find the correct magnitude. Nevertheless, lets get started.
try<-nls(Y ~ b0 + b1*sin(Position)+b2*log(Position),
data = df_long_group2,
start = list(b0 = 100,
b1 = 100,
b2 = 100),
na.action=na.exclude,
control=nls.control(maxiter = 1000, tol = 1e-05))
summary(try)
plot(try)
df_long_group2$pred<-predict(try)
ggplot(df_long_group2)+
geom_point(aes(x=Position,
y=Y))+
geom_line(aes(x=Position,
y=pred,
group=Condition), col="red")+
theme_bw()


Even though the model is not good, lets see how the Bayesian model performs using brms. I will apply the same function and part of the priors used in the book as well. These are some ver very uninformative priors. You can hardly call this Bayesian analysis.
fit2<-brms::brm(bf(Y | trials(N) ~
b0 + b1*sin(Position)+b2*log(Position),
b0 + b1 + b2 ~ 1 + (1|Condition), nl=TRUE),
data= df_long,
family=binomial(link = "logit"),
prior = c(
prior(uniform(0, 200), class=b, coef=Intercept, nlpar = "b0"),
prior(uniform(0, 200), class=b, coef=Intercept, nlpar = "b1"),
prior(uniform(0, 250), class=b, coef=Intercept, nlpar = "b2"),
prior(gamma(1, 1), class=sd, group=Condition, nlpar = "b0"),
prior(gamma(1, 1), class=sd, group=Condition, nlpar = "b1"),
prior(gamma(1, 1), class=sd, group=Condition, nlpar = "b2")),
chains=4,
cores=6,
warmup = 3000,
iter = 6000)
What will happen if I change the priors? I will now construct uniform priors for the location part and gamma priors for the scale part.
fit3<-brms::brm(bf(Y | trials(N) ~
b0 + b1*sin(Position)+b2*log(Position),
b0 + b1 + b2 ~ 1 + (1|Condition), nl=TRUE),
data= df_long,
family=binomial(link = "logit"),
prior = c(
prior(uniform(-10, 10), class=b, coef=Intercept, nlpar = "b0"),
prior(uniform(-10, 10), class=b, coef=Intercept, nlpar = "b1"),
prior(uniform(-10, 10), class=b, coef=Intercept, nlpar = "b2"),
prior(gamma(1, 1), class=sd, group=Condition, nlpar = "b0"),
prior(gamma(1, 1), class=sd, group=Condition, nlpar = "b1"),
prior(gamma(1, 1), class=sd, group=Condition, nlpar = "b2")),
chains=4,
cores=6,
warmup = 3000,
iter = 6000)
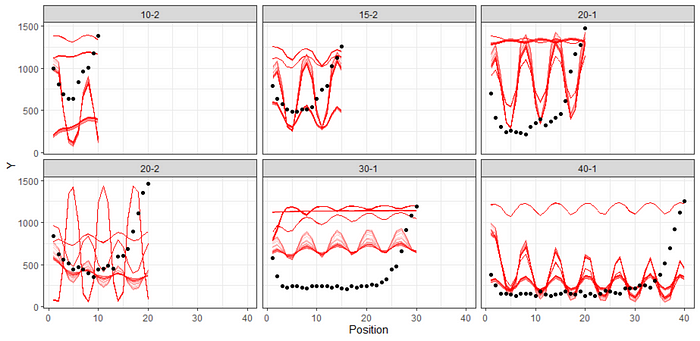
summary(fit3)
pp_check(fit3, ndraws=500)


The particular function used in the book is nested, and quite sophisticated. Of course, I could use the STAN code the authors applied, but at this point I do not even want to anymore. I want to see if I can model this pattern in a non-linear way. But before I do that, I want to take one last detour, and that is applying a polynomial function. Very much in line with a spline, it has some easier to interpret output. And its agile. Lets quickly assess.
First, I will test using a linear regression and the lm function.
df_long_group2<-df_long%>%filter(Condition=="20-1")
plot(df_long_group2$Position, df_long_group2$Y)
try<-lm(Y ~ Position +
I(Position^2) +
I(Position^3), data=df_long_group2)
summary(try)
df_long_group2$pred<-predict(try)
ggplot(df_long_group2)+
geom_point(aes(x=Position,
y=Y))+
geom_line(aes(x=Position,
y=pred,
group=Condition), col="red")+
theme_bw()

Now, I will try the same thing using the glm function.
try<-glm(cbind(Y|N) ~ Position +
I(Position^2) +
I(Position^3),
data=df_long_group2,
family=binomial)
summary(try)
df_long_group2$pred<-predict(try, df_long_group2, type="link")
ggplot(df_long_group2)+
geom_point(aes(x=Position,
y=Y))+
geom_line(aes(x=Position,
y=pred,
group=Condition), col="red")+
theme_bw()

In the end, I do not wish to model the data using a Normal distribution, but with the Binomial. For the priors I will take the normal for the location parameters and gamma for the scale / precision parameter. These are quite wide.
get_prior(bf(Y | trials(N) ~
Position +
I(Position^2) +
I(Position^3) +
(1|Condition)),
data= df_long,
family=binomial(link = "logit"))
fit4<-brms::brm(bf(Y | trials(N) ~
Position +
I(Position^2) +
I(Position^3) +
(1|Condition)),
data= df_long,
family=binomial(link = "logit"),
prior = c(
prior(normal(200, 50), class=Intercept),
prior(normal(0,10), class=b, coef=Position),
prior(normal(0,10), class=b, coef=IPositionE2),
prior(normal(0,10), class=b, coef=IPositionE3),
prior(gamma(4, 4), class=sd, group=Condition)),
chains=4,
cores=6,
warmup = 3000,
iter = 6000)
summary(fit4)
pp_check(fit4, ndraws=500)
df_long %>%
add_epred_draws(fit4,
ndraws=200,
allow_new_levels = TRUE) %>%
ggplot(aes(x = Position,
y = Y,
group=Condition)) +
geom_line(aes(y = .epred, group = paste(Condition, .draw)), alpha = 0.25, col="red") +
geom_point(data = df_long, color="black")+
facet_wrap(~Condition)+
theme_bw() 


Instead of the polynomial, I could revert back to splines, but this time I will ask the model to make splines per category. This is easily done in brms.
get_prior(bf(Y | trials(N) ~
s(Position, by=Condition)),
data= df_long,
family=binomial(link = "logit"))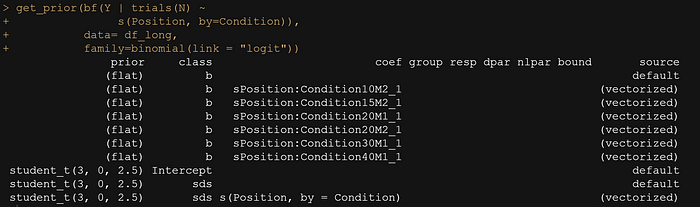
For this model, I will stick with uniform priors. Once again, this is weak Bayesian modelling, but I am unsuccessful using polynomials and so I wish to see where the data will bring me.
fit5<-brms::brm(bf(Y | trials(N) ~
s(Position, by=Condition)),
data= df_long,
family=binomial(link = "logit"),
prior = c(
prior(uniform(-10, 10), class=Intercept),
prior(uniform(-10, 10), class=b, coef=sPosition:Condition10M2_1),
prior(uniform(-10, 10), class=b, coef=sPosition:Condition15M2_1),
prior(uniform(-10, 10), class=b, coef=sPosition:Condition20M1_1),
prior(uniform(-10, 10), class=b, coef=sPosition:Condition20M2_1),
prior(uniform(-10, 10), class=b, coef=sPosition:Condition30M1_1),
prior(uniform(-10, 10), class=b, coef=sPosition:Condition40M1_1),
prior(gamma(1, 1), class=sds)),
chains=4,
cores=6,
warmup = 3000,
iter = 6000)
summary(fit5)
pp_check(fit5, ndraws=500)
df_long %>%
add_epred_draws(fit5,
ndraws=200,
allow_new_levels = TRUE) %>%
ggplot(aes(x = Position,
y = Y,
group=Condition)) +
geom_line(aes(y = .epred, group = paste(Condition, .draw)), alpha = 0.25, col="red") +
geom_point(data = df_long, color="black")+
facet_wrap(~Condition)+
theme_bw()

And the prediction, which do not look bad either. Remember, this is full statistical and not mechanical modelling. But in the end, the model does seem to do the trick, at the expense of some loss of information on what the parameters exactly mean.

I am still not satisfied and wish to take a look at a non-linear function one last time. Lets go at it again.
df_long_group2<-df_long%>%filter(Condition=="20-1")
plot(df_long_group2$Position, df_long_group2$Y)
try<-nls(Y ~ a1*exp(b1*Position)+c1,
data = df_long_group2,
start = list(a1 = 1,
b1 = 1,
c1 = 1),
na.action=na.exclude,
control=nls.control(maxiter = 1000, tol = 1e-05))
summary(try)
df_long_group2$pred<-predict(try)
ggplot(df_long_group2)+
geom_point(aes(x=Position,
y=Y))+
geom_line(aes(x=Position,
y=pred,
group=Condition), col="red")+
theme_bw()

Lets try the same non-linear function on another condition.
df_long_group2<-df_long%>%filter(Condition=="40-1")
plot(df_long_group2$Position, df_long_group2$Y)
try<-nls(Y ~ a1*exp(b1*Position)+c1,
data = df_long_group2,
start = list(a1 = 2,
b1 = 0.5,
c1 = 300),
na.action=na.exclude,
control=nls.control(maxiter = 1000, tol = 1e-05))
summary(try)
df_long_group2$pred<-predict(try)
ggplot(df_long_group2)+
geom_point(aes(x=Position,
y=Y))+
geom_line(aes(x=Position,
y=pred,
group=Condition), col="red")+
theme_bw()

On to the next challenge, which is to place the non-linear function in the Bayesian framework.
get_prior(bf(Y | trials(N) ~
a1*exp(b1*Position)+c1,
a1 + b1 + c1 ~ 1, nl=TRUE),
data= df_long,
family=binomial(link = "logit"))
fit7<-brms::brm(bf(Y | trials(N) ~
a1*exp(b1*Position)+c1,
a1 + b1 + c1 ~ 1, nl=TRUE),
data= df_long,
family=binomial(link = "logit"),
prior = c(
prior(uniform(-10, 10), class=b, coef=Intercept, nlpar = "a1"),
prior(uniform(-10, 10), class=b, coef=Intercept, nlpar = "b1"),
prior(uniform(-400, 400), class=b, coef=Intercept, nlpar = "c1")),
chains=4,
cores=6,
warmup = 3000,
iter = 6000)
summary(fit7)
df_long %>%
add_epred_draws(fit7,
ndraws=100,
allow_new_levels = TRUE) %>%
ggplot(aes(x = Position,
y = Y,
group=Condition)) +
geom_line(aes(y = .epred, group = paste(Condition, .draw)), alpha = 0.25, col="red") +
geom_point(data = df_long, color="black")+
facet_wrap(~Condition)+
theme_bw() 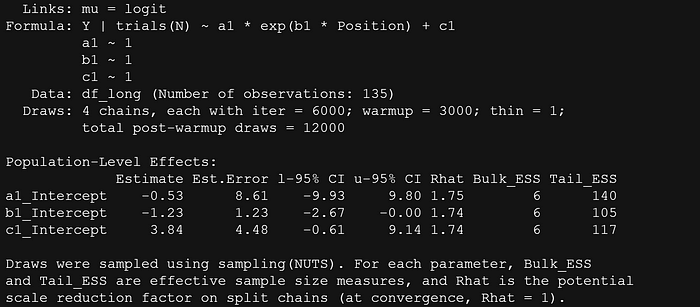
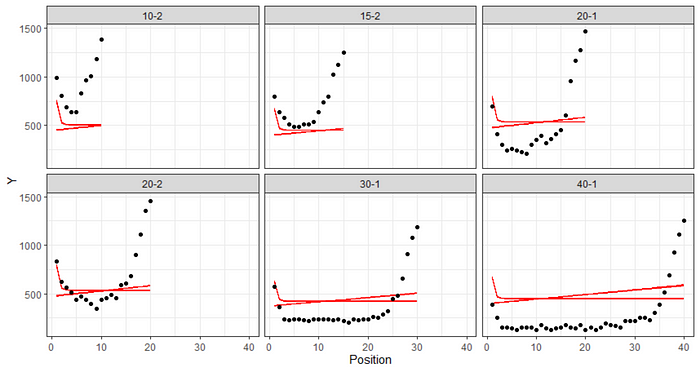
A next try, which will not add much, but I am still doing it, is to build a full non-linear mixed model in which the three parameters are allowed to shift per condition. For the location parameters I am using a uniform prior, and for the scale I am using gamma.
fit7<-brms::brm(bf(Y | trials(N) ~
a1*exp(b1*Position)+c1,
a1 + b1 + c1 ~ 1 + (1|Condition), nl=TRUE),
data= df_long,
family=binomial(link = "logit"),
prior = c(
prior(uniform(-10, 10), class=b, coef=Intercept, nlpar = "a1"),
prior(uniform(-10, 10), class=b, coef=Intercept, nlpar = "b1"),
prior(uniform(-300, 300), class=b, coef=Intercept, nlpar = "c1"),
prior(gamma(1, 1), class=sd, group=Condition, nlpar = "a1"),
prior(gamma(1, 1), class=sd, group=Condition, nlpar = "b1"),
prior(gamma(1, 1), class=sd, group=Condition, nlpar = "c1")),
chains=4,
cores=6,
warmup = 3000,
iter = 6000)
summary(fit7)
df_long %>%
add_epred_draws(fit7,
ndraws=100,
allow_new_levels = TRUE) %>%
ggplot(aes(x = Position,
y = Y,
group=Condition)) +
geom_line(aes(y = .epred, group = paste(Condition, .draw)), alpha = 0.25) +
geom_point(data = df_long, color="black")+
facet_wrap(~Condition)+
theme_bw()

Perhaps with some more informative priors, or adding a parameter to the non-linear function I could have improved the model, but for now this is what it is.
In the end, I will stick with the conditional spline model, which builds a spline function per condition. This is not exactly the same model as the one in the book, but it is a model nonetheless.
For my next series, I will look deeper in STAN function itself, which will offer much more versatility.
If anything is amiss, or you just have a comment, let me know!

